Week III: Biological Databases & Accessing databases¶
Summary¶
In the realm of modern genomic research, a notable characteristic is the production of vast quantities of raw sequence data. As the volume of genomic information expands, the management of this data surge requires advanced computational methods. Thus, the initial challenge in genomics lies in efficiently storing and handling this immense amount of data through the establishment and utilization of computer databases. Developing databases capable of managing the extensive molecular biological data is a fundamental task in the field of bioinformatics. This chapter provides an introduction to fundamental concepts related to databases, particularly focusing on the types, structures, and architectures of biological databases. It places emphasis on data retrieval from major biological databases like GenBank.
What is database?¶
A database serves as a computerized repository designed to store and structure data in a manner that facilitates easy retrieval through various search criteria. Databases consist of both computer hardware and software components dedicated to data management. The primary goal in creating a database is to arrange data into structured records, allowing for straightforward retrieval of information. Each record, also referred to as an entry, should include multiple fields for holding specific data items, such as fields for names, phone numbers, addresses, and dates. When seeking a specific record within the database, a user can specify a particular piece of information, called a value, located in a specific field, and expect the computer to retrieve the entire data record.
Biological Databases¶
Biological databases can be categorized into three main types according to their content: primary databases, secondary databases, and specialized databases.
Primary databases store original biological data, serving as repositories for the unprocessed raw sequence or structural data contributed by the scientific community. Notable examples of primary databases include GenBank and the Protein Data Bank (PDB).
Secondary databases contain information that has been computationally processed or manually curated. This information is derived from the original data found in primary databases. This category includes databases housing translated protein sequences with functional annotations such as UniProt.
Specialized databases are tailored to meet specific research interests. They are designed to focus on particular organisms or specific types of data. For instance, databases like Flybase, the HIV sequence database, and the Ribosomal Database Project are examples of specialized databases, each specializing in a particular organism or a specific data type. Another example NCBI PubMed database which is a biomedical literature database supporting the search and retrieval of biomedical and life sciences literature. PubMed contains more than 36 million citations and abstracts of biomedical literature.
About the Content
Citations in PubMed primarily stem from the biomedicine and health fields, and related disciplines such as life sciences, behavioral sciences, chemical sciences, and bioengineering.
Nucleotide Sequence Databases¶
GenBank¶
GenBank is the NCBI genetic sequence database, an annotated collection of all publicly available DNA sequences. GenBank is part of the International Nucleotide Sequence Database Collaboration, which comprises the DNA DataBank of Japan (DDBJ), the European Nucleotide Archive (ENA), and GenBank at NCBI. These three organizations exchange data on a daily basis. GenBank releases occur every two months and is available from the ftp site.
DDBJ¶
DDBJ (DNA Data Bank of Japan) shares annotated/assembled nucleotide sequence data as a member of INSDC (International Nucleotide Sequence Database Collaboration). DDBJ Center provides freely available nucleotide sequence data and supercomputer system, to support research activities in life science.
EMBL ENA¶
The European Nucleotide Archive (ENA) provides a comprehensive record of the world’s nucleotide sequencing information, covering raw sequencing data, sequence assembly information and functional annotation. More about ENA.
miRBase¶
miRBase is a biological database that acts as an archive of microRNA sequences and annotations.
Rfam¶
Rfam is a database containing information about non-coding RNA (ncRNA) families and other structured RNA elements.
Genome Databases¶
NCBI Genome Database¶
The Genome database organizes information on genomes including sequences, maps, chromosomes, assemblies, and annotations. This resource contains sequence and map data from the whole genomes of over 1000 species or strains. The genomes represent both completely sequenced genomes and those with sequencing in-progress. All three main domains of life (bacteria, archaea, and eukaryota) are represented, as well as many viruses, phages, viroids, plasmids, and organelles.
Ensembl¶
Ensembl is a genome browser for vertebrate genomes that supports research in comparative genomics, evolution, sequence variation and transcriptional regulation. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data. Ensembl tools include BLAST, BLAT, BioMart and the Variant Effect Predictor (VEP) for all supported species.
Mostly gene identifiers (gene IDs) of expression data are Ensembl Gene IDs and they need to be converted to different gene IDs or symbols of interest using BioMart tool.
GOLD¶
GOLD (Genomes Online Database), is a resource for comprehensive access to information regarding genome and metagenome sequencing projects, and their associated metadata, around the world.
Domain and Motif Databases¶
InterPro¶
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites. To classify proteins in this way, InterPro uses predictive models, known as signatures, provided by several different databases (referred to as member databases) that make up the InterPro consortium. InterPro combines protein signatures from these member databases into a single searchable resource, capitalising on their individual strengths to produce a powerful integrated database and diagnostic tool.
Prosite¶
PROSITE is a database of protein families and domains. It is based on the observation that, while there is a huge number of different proteins, most of them can be grouped, on the basis of similarities in their sequences, into a limited number of families. Proteins or protein domains belonging to a particular family generally share functional attributes and are derived from a common ancestor. PROSITE is complemented by ProRule, a collection of rules based on profiles and patterns, which increases the discriminatory power of profiles and patterns by providing additional information about functionally and/or structurally critical amino acids.
CDD¶
Conserved Domain Database (CDD) is a protein annotation resource that consists of a collection of well-annotated multiple sequence alignment models for ancient domains and full-length proteins. These are available as position-specific score matrices (PSSMs) for fast identification of conserved domains in protein sequences via RPS-BLAST. CDD content includes NCBI-curated domains, which use 3D-structure information to explicitly define domain boundaries and provide insights into sequence/structure/function relationships, as well as domain models imported from a number of external source databases (Pfam, SMART, COG, PRK, TIGRFAMs).
SMART¶
SMART (Simple Modular Architecture Research Tool) allows the identification and annotation of genetically mobile domains and the analysis of domain architectures. More than 500 domain families found in signalling, extracellular and chromatin-associated proteins are detectable. These domains are extensively annotated with respect to phyletic distributions, functional class, tertiary structures and functionally important residues. Each domain found in a non-redundant protein database as well as search parameters and taxonomic information are stored in a relational database system.
COG¶
COGs (Clusters of Orthologous Genes) database includes complete genomes of 1,187 bacteria and 122 archaea that map into 1,234 genera. The new features include ~250 updated COG annotations with corresponding references and PDB links, where available; new COGs for proteins involved in CRISPR-Cas immunity, sporulation, and photosynthesis, and the lists of COGs grouped by pathways and functional systems.
TOPDOM¶
TOPDOM (Conservatively Located Domains and Motifs in Proteins) is a collection of domains and sequence motifs located conservatively on one side of membranes either in transmembrane or globular proteins. The database was created by predicting the transmembrane status and topology of all protein sequences in UniProt (SwissProt) database by the CCTOP algorithm and scanning by specific domain or motif detecting algorithms. The identified domain or motif was added to the database if it was uniformly annotated in the same side of the membrane of the various proteins in UniProt (SwissProt) database. The sequences in the UniProt (SwissProt) database were scanned by InterProScan algorithm for domains and motifs from Gene3D, TIGRFAM/NCBIfam, PANTHER, PRINTS, Pfam, ProSiteProfiles, SMART, SUPERFAMILY.
Protein Sequence Databases¶
UniProt¶
The Universal Protein Resource (UniProt) is the world’s leading high-quality, comprehensive and freely accessible resource of protein sequence and functional information. UniProt is a comprehensive resource for protein sequence and annotation data. The UniProt databases are the UniProt Knowledgebase (UniProtKB), the UniProt Reference Clusters (UniRef), and the UniProt Archive (UniParc). The Protein Information Resource (PIR), along with its international partners, EBI (European Bioinformatics Institute) and SIB (Swiss Institute of Bioinformatics), were awarded a grant from NIH to create UniProt, a single worldwide database of protein sequence and function, by unifying the PIR-PSD, Swiss-Prot, and TrEMBL databases.
NCBI Protein Database¶
The Protein database is a collection of sequences from several sources, including translations from annotated coding regions in GenBank, RefSeq and TPA, as well as records from SwissProt, PIR, PRF, and PDB. Protein sequences are the fundamental determinants of biological structure and function.
Protein Structure Databases¶
wwPDB¶
The Protein Data Bank archive (PDB) has served as the single repository of information about the 3D structures of proteins, nucleic acids, and complex assemblies.
wwPDB Members
Research Collaboratory for Structural Bioinformatics Protein Data Bank
Simple and advanced searching for macromolecules and ligands, flexible APIs for programmatic users, specialized 1D-3D visualization tools, Molecule of the Month and other training resources at PDB-101, and more.
Protein Data Bank in Europe
Rich information about all PDB entries, multiple search and browse facilities, advanced services including PDBePISA, PDBeFold and PDBeMotif, advanced visualisation and validation of NMR and EM structures, tools for bioinformaticians.
Protein Data Bank Japan
Supports browsing in multiple languages such as Japanese, Chinese, and Korean; SeSAW identifies functionally or evolutionarily conserved motifs by locating and annotating sequence and structural similarities, tools for bioinformaticians, and more.
Electron Microscopy Data Bank
Collects 3D volumes & associated information of macromolecular complexes & subcellular structures from electron cryo microscopy & electron cryo tomography; develops resources for searching, data mining, analyzing, validating & visualizing data.
Biological Magnetic Resonance Data Bank
Collects NMR data from any experiment and captures assigned chemical shifts, coupling constants, and peak lists for a variety of macromolecules; contains derived annotations such as hydrogen exchange rates, pKa values, and relaxation parameters.
NCBI Structure¶
The Molecular Modeling Database (MMDB), as part of the Entrez system, facilitates access to structure data by connecting them with associated literature, protein and nucleic acid sequences, chemicals, biomolecular interactions, and more.
SCOP¶
SCOP (Structural Classification of Proteins) database, created by manual inspection and abetted by a battery of automated methods, aims to provide a detailed and comprehensive description of the structural and evolutionary relationships between all proteins whose structure is known. As such, it provides a broad survey of all known protein folds, detailed information about the close relatives of any particular protein, and a framework for future research and classification.
CATH¶
The CATH database is a free, publicly available online resource that provides information on the evolutionary relationships of protein domains. Experimentally-determined protein three-dimensional structures are obtained from the Protein Data Bank and split into their consecutive polypeptide chains. Protein domains are identified within these chains using a mixture of automatic methods and manual curation. The domains are then classified within the CATH structural hierarchy.
AlphaFold Protein Structure Database¶
Gene Expression Databases¶
GEO¶
GEO is an international public repository that archives and freely distributes microarray, next-generation sequencing, and other forms of high-throughput functional genomics data submitted by the research community. The GEO DataSets stores original submitter-supplied records (Series, Samples and Platforms) as well as curated DataSets.
GXD¶
MGI-Mouse Gene Expression Database (GXD) is a community resource for gene expression information from the laboratory mouse. GXD stores and integrates different types of expression data and makes these data freely available in formats appropriate for comprehensive analysis. There is particular emphasis on endogenous gene expression during mouse development.
ArrayExpress¶
The functional genomics data collection (ArrayExpress), stores data from high-throughput functional genomics experiments, and provides data for reuse to the research community. In line with community guidelines, a study typically contains metadata such as detailed sample annotations, protocols, processed data and raw data. Raw sequence reads from high-throughput sequencing studies are brokered to the European Nucleotide Archive (ENA), and links are provided to download the sequence reads from ENA. Data can be submitted to the ArrayExpress collection through its dedicated submission tool, Annotare.
Pathway Databases¶
KEGG¶
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies.
Reactome¶
Reactome is a pathway database containing cell metabolic and signaling pathways. Cold Spring Harbor Laboratory, European Bioinformatics Institute, and Gene Ontology Consortium are the main developers of the project. It has data for 23 species such as human, mouse and rat. The storage format is proprietary, a large number of pathways can be obtained in multiple formats.
BioCyc¶
BioCyc is a pathway database focused on metabolic pathways originally formed by SRI International’s bioinformatics research group. Related to BioCyc are the EcoCyc (E. coli Database), MetaCyc (Metabolic Pathway Database), HumanCyc (Encyclopedia of Human Genes and Metabolism) databases. Humans and E. coli are the major organisms listed with a variety of others.
WikiPathways¶
WikiPathways is an open, collaborative platform dedicated to the curation of biological pathways.
Pathway Commons¶
Pathway Commons collects and disseminates biological pathway and interaction data.
Pathguide¶
Pathguide contains information about 702 biological pathway related resources and molecular interaction related resources.
Human MSigDB¶
Human MSigDB is the Human Molecular Signatures Database (MSigDB) consisting of the 33591 gene sets divided into 9 major collections, and several subcollections.
miRPathDB¶
miRPathDB is one of the most comprehensive and advanced resources for miRNAs and their target pathways.
BRENDA¶
BRENDA is the main collection of enzyme functional data available to the scientific community.
Specialised Databases¶
PhosphoSitePlus¶
PhosphoSitePlus provides comprehensive information and tools for the study of protein post-translational modifications (PTMs) including phosphorylation, acetylation, and more.
GPCRD¶
The GPCRdb contains data, diagrams and web tools for G protein-coupled receptors (GPCRs).
Databases for various model organisms¶
NCBI GenBank Record Format¶
Each database stores the information with unified database IDs. For each sequence the NCBI Nucleotide database stores records with accession number as ID, NCBI GenBank stores records with Gene ID and some extra information such as the species that it came from, publications describing the sequence, etc.
To view the GenBank entry for the DEN-1 Dengue virus, follow these steps:
- Go to the NCBI website (www.ncbi.nlm.nih.gov).
- Search for the accession number NC_001477.
- Since we searched for a particular accession we are only returned a single main result which is titled “NUCLEOTIDE SEQUENCE: Dengue virus 1, complete genome.”
- Click on “Dengue virus 1, complete genome” to go to the GenBank entry. The GenBank entry for an accession contains a lot of information about the sequence, such as papers describing it, features in the sequence, etc. The DEFINITION field gives a short description for the sequence. The ORGANISM field in the NCBI entry identifies the species that the sequence came from. The REFERENCE field contains scientific publications describing the sequence. The FEATURES field contains information about the location of features of interest inside the sequence, such as regulatory sequences or genes that lie inside the sequence. The ORIGIN field gives the sequence itself.
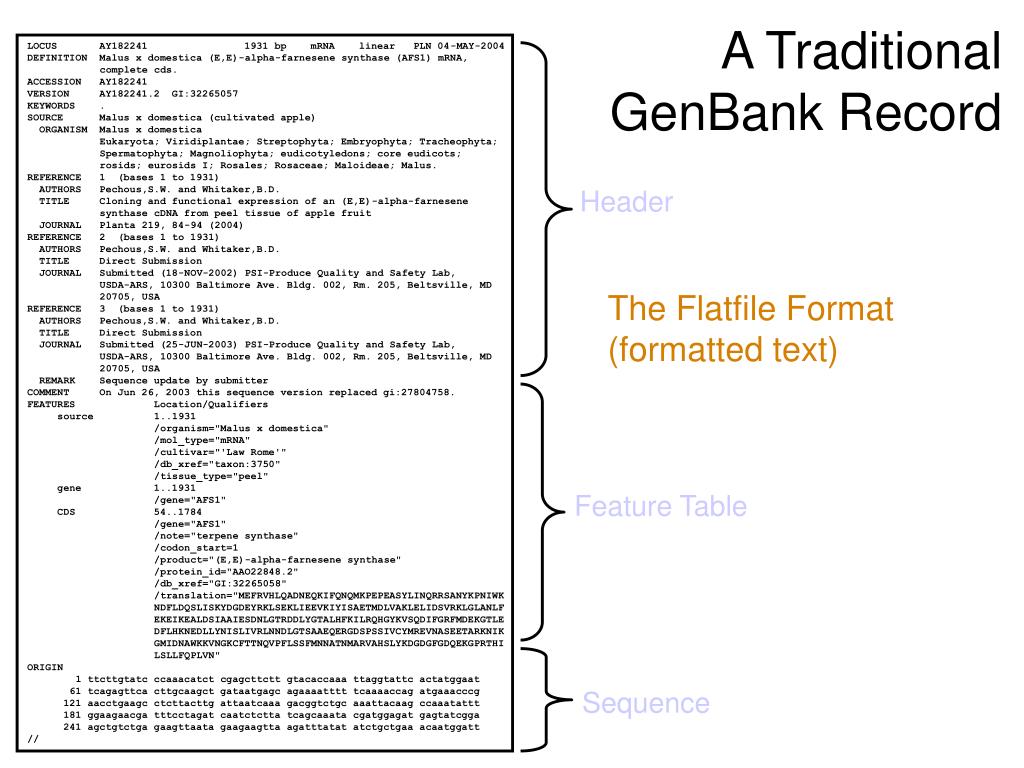
Figure 1. A typical NCBI Nucleotide database record.
This page presents an annotated sample GenBank record (accession number U49845) in its GenBank Flat File format. You can find explanation of the information fields.
The FASTA file format¶
The FASTA file format is a simple file format commonly used to store and share sequence information. When you download sequences from databases such as NCBI you usually want FASTA files.
The first line of a FASTA file starts with the “greater than” character (>) followed by a name and/or description for the sequence. Subsequent lines contain the sequence itself. A short FASTA file might contain just something like this:
>U49845.1 Saccharomyces cerevisiae TCP1-beta gene, partial cds; and Axl2p (AXL2) and Rev7p (REV7) genes, complete cds
GATCCTCCATATACAACGGTATCTCCACCTCAGGTTTAGATCTCAACAACGGAACCATTGCCGACATGAG
ACAGTTAGGTATCGTCGAGAGTTACAAGCTAAAACGAGCAGTAGTCAGCTCTGCATCTGAAGCCGCTGAA
A FASTA file can contain the sequence for a single, an entire genome, or more than one sequence. If a FASTA file contains many sequences, then for each sequence it will have a header line starting with a greater than character followed by the sequence itself.
This is what a FASTA file with two sequence looks like.
>mysequence1
GATCCTCCATATACAACGGTATCTCCACCTCAGGTTTAGATCTCAACAACGGAACCATTGCCGACATGAG
ACAGTTAGGTATCGTCGAGAGTTACAAGCTAAAACGAGCAGTAGTCAGCTCTGCATCTGAAGCCGCTGAA
>mysequence2
GTTCTACTAAGGGTGGATAACATCATCCGTGCAAGACCAAGAACCGCCAATAGACAACATATGTAACATA
TTTAGGATATACCTCGAAAATAATAAACCGCCACACTGTCATTATTATAATTAGAAACAGAACGCAAAAA
TTATCCACTATATAATTCAAAGACGCGAAAAAAAAA
Querying the NCBI Database¶
You may need to interrogate the NCBI Database to find particular sequences or a set of sequences matching given criteria, such as:
The sequence with accession NC_001477 All sequences from Chlamydia trachomatis Flagellin or fibrinogen sequences The glutamine synthetase gene from Mycobacteriuma leprae Just the upstream control region of the Mycobacterium leprae dnaA gene The sequence of the Mycobacterium leprae DnaA protein The genome sequence of syphilis, Treponema pallidum subspp. pallidum All human nucleotide sequences associated with malaria
There are two main ways that you can query the NCBI database to find these sets of sequences. The first possibility is to carry out searches on the NCBI website. The second possibility is to carry out searches from R or Python packages that can interface with NCBI.
Querying the NCBI Database via the NCBI Website:
If you are carrying out searches on the NCBI website, to narrow down your searches to specific types of sequences or to specific organisms, you will need to use “search tags”.
For example, the search tags “[PROP]” and “[ORGN]” let you restrict your search to a specific subset of the NCBI Sequence Database, or to sequences from a particular taxon, respectively. Here is a list of useful search tags, which we will explain how to use below:
- [AC], e.g. NC_001477[AC] With a particular accession number
- [ORGN], e.g. Fungi[ORGN] From a particular organism or taxon
- [PROP], e.g. biomol_mRNA[PROP] Of a specific type (eg. mRNA) or from a specific database (eg. RefSeq)
To carry out searches of the NCBI database, you first need to go to the NCBI website, and type your search query into the search box at the top. For example, to search for all sequences from Fungi, you would type “Fungi[ORGN]” into the search box on the NCBI website.
You can combine the search tags above by using “AND”, to make more complex searches. For example, to find all mRNA sequences from Fungi, you could type “Fungi[ORGN] AND biomol_mRNA[PROP]” in the search box on the NCBI website.
Likewise, you can also combine search tags by using “OR”, for example, to search for all mRNA sequences from Fungi or Bacteria, you would type “(Fungi[ORGN] OR Bacteria[ORGN]) AND biomol_mRNA[PROP]” in the search box. Note that you need to put brackets around “Fungi[ORGN] OR Bacteria[ORGN]” to specify that the word “OR” refers to these two search tags.
Here are some examples of searches, some of them made by combining search terms using “AND”:
- NC_001477[AC] - With accession number NC_001477
- “Chlamydia trachomatis”[ORGN] - From the bacterium Chlamydia trachomatis
- flagellin OR fibrinogen - Which contain the word “flagellin” or “fibrinogen” in their NCBI record
- “Mycobacterium leprae”[ORGN] AND dnaA - Which are from M. leprae, and contain “dnaA” in their NCBI record
- “Homo sapiens”[ORGN] AND “colon cancer” - Which are from human, and contain “colon cancer” in their NCBI record
- “Homo sapiens”[ORGN] AND malaria - Which are from human, and contain “malaria” in their NCBI record
- “Homo sapiens”[ORGN] AND biomol_mrna[PROP] - Which are mRNA sequences from human
- “Bacteria”[ORGN] AND srcdb_refseq[PROP] - Which are RefSeq sequences from Bacteria
- “colon cancer” AND srcdb_refseq[PROP] - From RefSeq, which contain “colon cancer” in their NCBI record
Note that if you are searching for a phrase such as “colon cancer” or “Chlamydia trachomatis”, you need to put the phrase in quotes when typing it into the search box. This is because if you type the phrase in the search box without quotes, the search will be for NCBI records that contain either of the two words “colon” or “cancer” (or either of the two words “Chlamydia” or “trachomatis”), not necessarily both words.
NCBI database contains several sub-databases, including the NCBI Nucleotide database and the NCBI Protein database. If you go to the NCBI website, and type one of the search queries above in the search box at the top of the page, the results page will tell you how many matching NCBI records were found in each of the NCBI sub-databases.
For example, if you search for “Chlamydia trachomatis[ORGN]”, you will get matches to proteins from C. trachomatis in the NCBI Protein database, matches to DNA and RNA sequences from C. trachomatis in the NCBI Nucleotide database, matches to whole genome sequences for C. trachomatis strains in the NCBI Genome database, and so on:
Alternatively, if you know in advance that you want to search a particular sub-database, for example, the NCBI Protein database, when you go to the NCBI website, you can select that sub-database from the drop-down list above the search box, so that you will search that sub-database.
Downloading genome data from NCBI¶
Downloading genome data from NCBI with Biopython and Entrez
View Jupyter Notebook